Methods: Philips et al 2020 “Thalamoseq”
Cell-types: Neurons
Brain regions: Thalamus
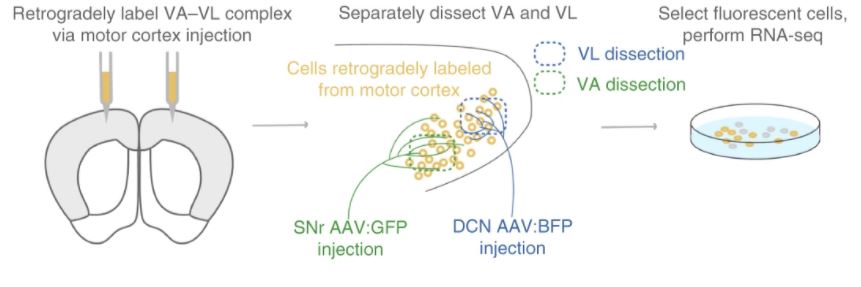
Techniques: Pooled cell RNAseq of thalamic nuclei: Retrograde labeling; anterograde tracing of thalamic inputs; manual cell-sorting; RNA-seq (pooled neurons) with HiSeq 2500. Single-cell RNAseq: Retrograde labeling; manual cell-sorting to 8-well strip; RNAseq with NextSeq 550. Single-cells (477 for motor, 282 for somatosensory, 429 for visual, 376 for auditory and 388 for prefrontal) were assigned to 17 cell-type (metacell) clusters (e.g. Aud_1).
Details of mice: C57Bl/6 background; 8-12 weeks
Data processing for BrainPalmSeq: Single-cell and nuclear data were downloaded from GEO with accession ID GSE133912 and GSE133911, respectively (count files GSE133912_thal_singlecell_counts.csv and GSE133911_thal_nuclei_counts.csv; annotation files GSE133912_thal_singlecell_metadata.csv and GSE133911_thal_nuclei_metadata.csv). Counts were log normalized by first scaling the expression values provided to a sum of 10,000 per cell before calculating log2(scaled_counts+1). Averages were then performed by cluster and projection for each gene for the single cell dataset, and by cluster and region for the nuclear dataset. Genes associated with palmitoylation were selected in order to create the heatmaps, categories comprising fewer than 5 single cells are not displayed.
Heatmap units: mean log2(counts per 10,000 + 1)
References: Phillips, J.W., Schulmann, A., Hara, E., Winnubst, J., Liu, C., Valakh, V., Wang, L., Shields, B.C., Korff, W., Chandrashekar, J., et al. (2019). A repeated molecular architecture across thalamic pathways. Nature Neuroscience 22, 1925–1935.
Website: https://thalamoseq.janelia.org/
Original data availability: GSE133911 (pooled-cell); GSE133912 (single-cell)