Methods: Saunders et al 2018 “DropViz”
Cell-types: All
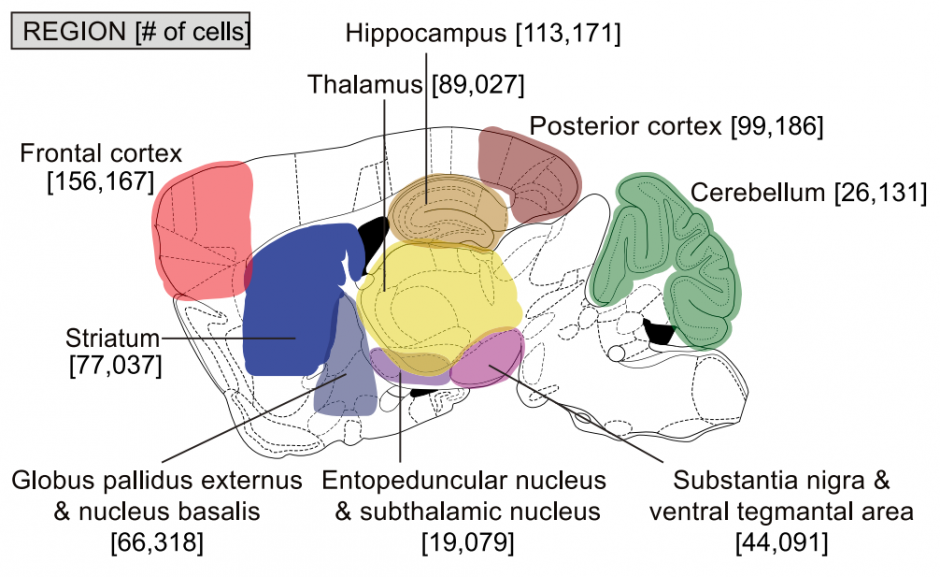
Brain regions: Frontal cortex; Posterior cortex; Striatum; Globus pallidus externus & nucleus basalis; Entopeduncular nucleus & subthalamic nucleus; Thalamus; Hippocampus; Substantia nigra & ventral tegmental area; Cerebellum.
Techniques: Selection-free cell-sampling of dissected brain regions. Single-cell RNAseq using Drop-seq protocol v3.1. 690,000 cells were assigned to 565 cell-type (metacell) clusters (e.g. CB_1-1).
Age of mice: P60-70
Data processing for BrainPalmSeq: Metacell counts were downloaded from DropViz.org (count file metacells.BrainCellAtlas_Saunders_version_2018.04.01.RDS and annotation file annotation.BrainCellAtlas_Saunders_version_2018.04.01.RDS) and log normalized by first scaling the expression values provided to a sum of 10,000 per metacell before calculating log2(scaled_counts+1). Averages were then performed by cell type, tissue and class for each gene. Genes associated with palmitoylation were selected in order to create the heatmaps.
Heatmap units: mean log2(counts per 10,000 + 1)
References: Saunders, A., Macosko, E.Z., Wysoker, A., Goldman, M., Krienen, F.M., de Rivera, H., Bien, E., Baum, M., Bortolin, L., Wang, S., et al. (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015-1030.e16.
Website: http://dropviz.org/
Original data availability: http://dropviz.org/ (Data)